基因分析在頭頸癌治療中的奇蹟:個體化醫療的新里程碑
基因分析在頭頸癌治療中的奇蹟:個體化醫療的新里程碑
隨著科技的進步,個人化基因分析正成為癌症治療領域的一項革命性技術。這種分析方法主要關注於檢測個體體細胞中的基因變異,以更準確地選擇適合的藥物,提高治療效果,同時預測疾病風險。今天,我們將聚焦在頭頸部鳞狀上皮細胞癌患者中,個人化基因分析是如何發揮作用,以及其在治療和風險預測方面的關鍵角色。
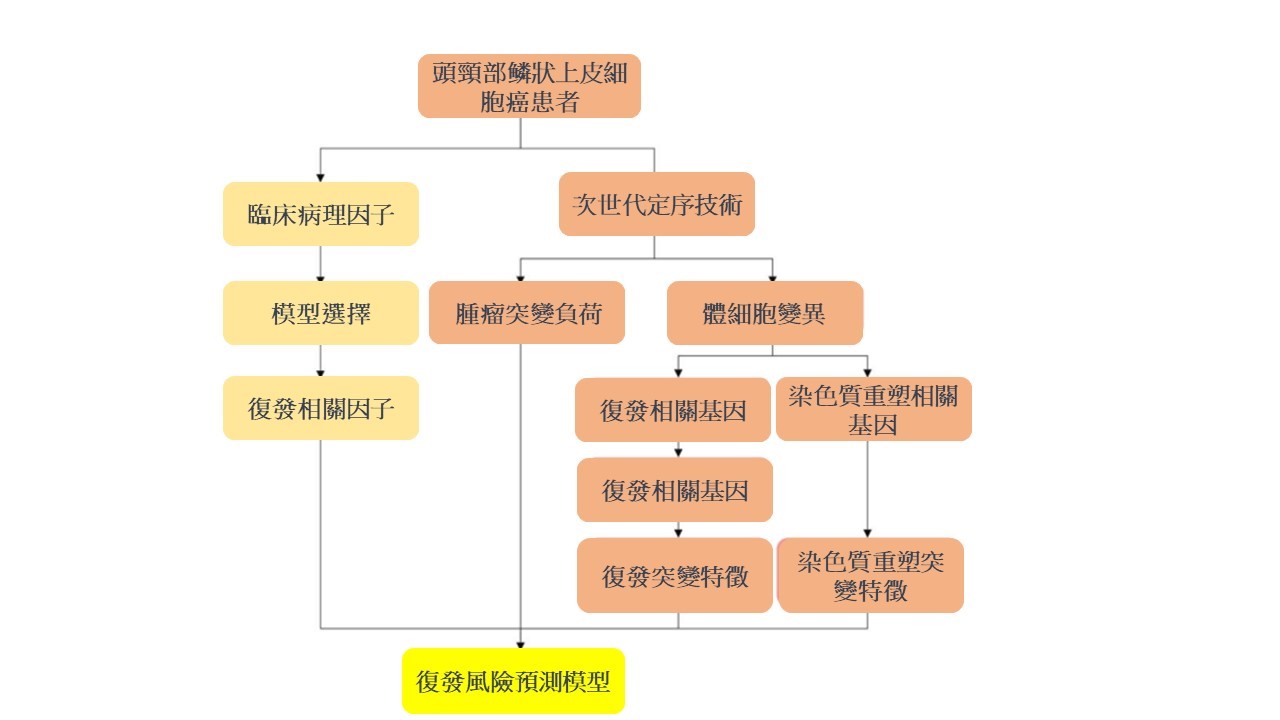
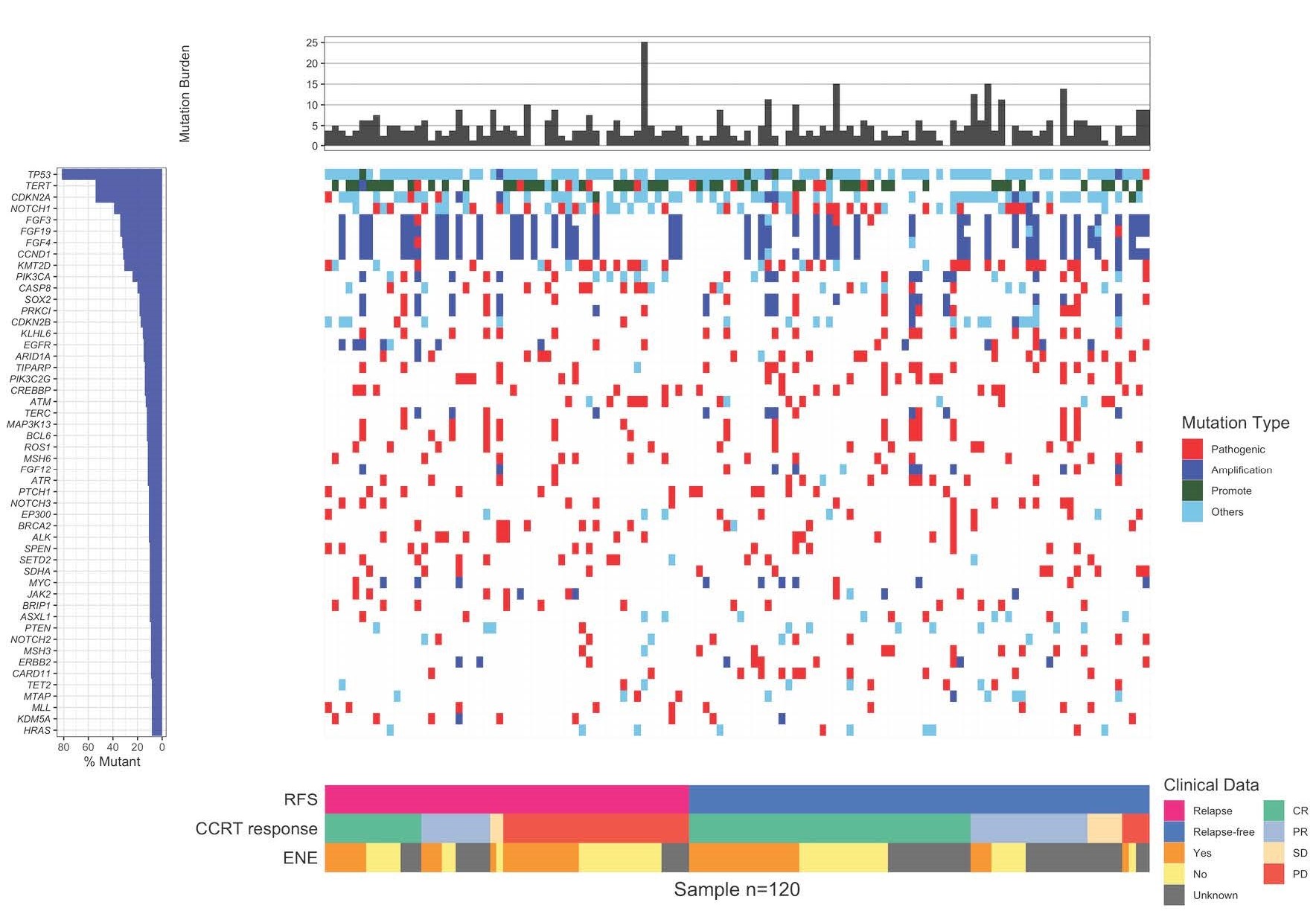
首先,讓我們了解個人化基因分析的基本原理。這項技術利用次世代定序技術,通過分析個體基因組中的突變,特別是腫瘤細胞中的變異,來確定最有效的治療方法。在頭頸癌的案例中,研究人員發現,這種分析可以區分出患者的復發風險,這對於預防和提早治療復發至關重要。
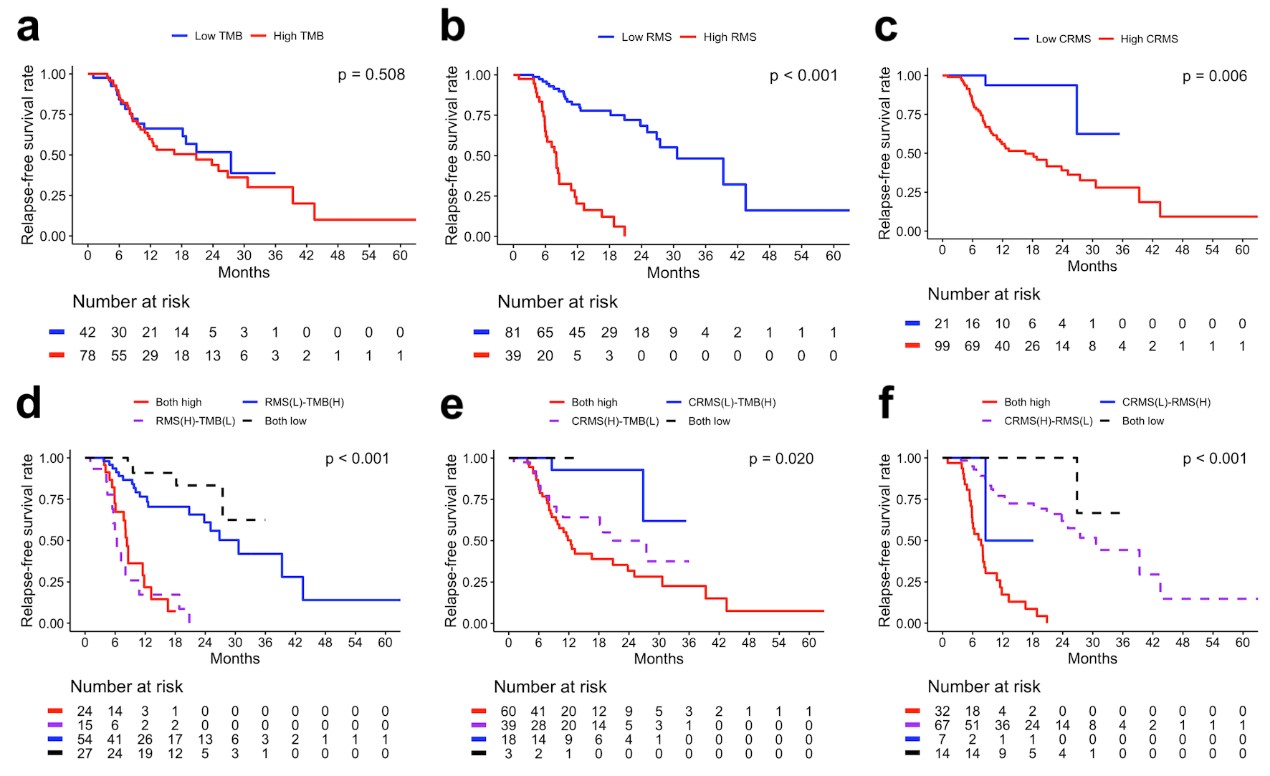
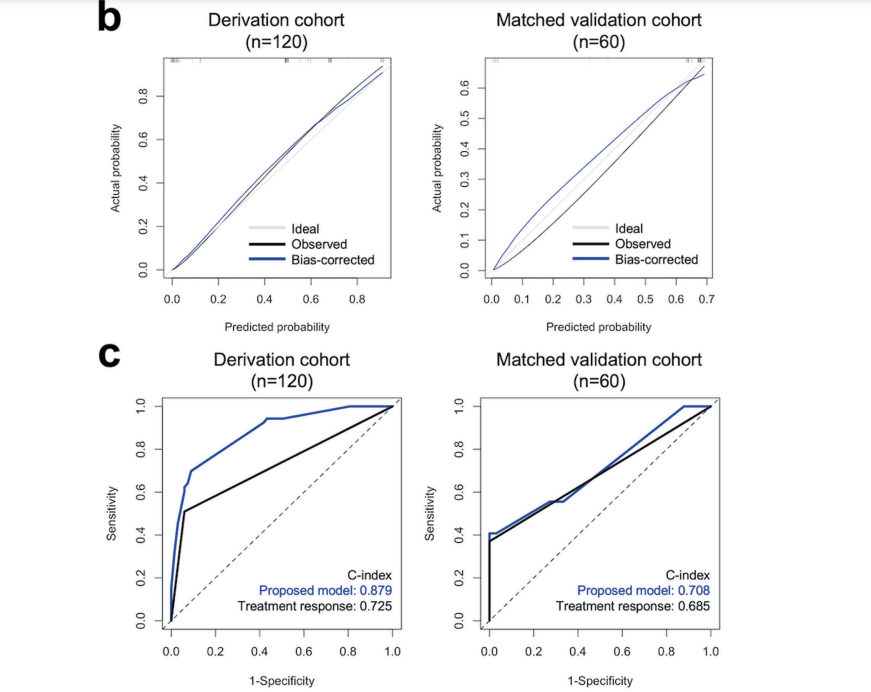
研究人員在探索基因分析和腫瘤突變負荷(TMB)的作用時,發現了復發突變特徵(RMS)和染色質重塑突變特徵(CRMS)這兩個關鍵指標。這兩個特徵的存在可以預測頭頸部鳞狀上皮細胞癌患者在接受鉑金藥物、同步放射線和化學治療後的復發風險。簡單來說,這項研究發現高RMS和CRMS組的患者相較於低RMS和CRMS組的患者,有更短的無復發生存期,這是一個極具臨床意義的發現(p < 0.001和p = 0.006)。
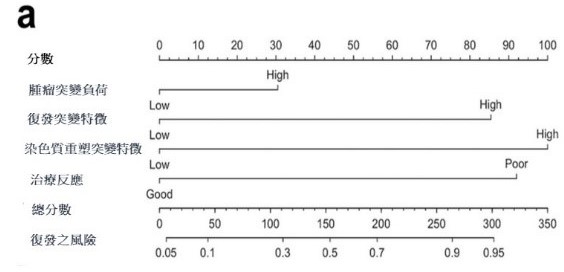
進一步的多變數Cox回歸分析強調了幾個獨立風險預測因子,包括淋巴結外侵犯、同步放射線和化學治療的反應,以及三種基因變異情形(TMB、RMS和CRMS)。這些因子組合成了一個預測性診斷模型,該模型在預測接受同步放射線和化學治療的頭頸部鳞狀上皮細胞癌患者的無復發生存期方面表現良好。
這項研究突顯了個人化基因分析在癌症治療中的潛在優勢。通過深入了解患者的基因組特徵,我們能夠更準確地預測治療效果和疾病風險,為每位患者提供更加個體化、精準的醫療方案。這一新里程碑將為未來的癌症治療開啟嶄新的可能性,帶來更多的希望和治療選擇。
圖形摘要
應用與亮點:
1.利用次世代定序技術進行基因分析和 TMB 的作用: 使用先進的定序技術評估基因分析和腫瘤突變負荷
(TMB),有助於更全面地了解患者的遺傳變異情況。
2.預測復發風險的突變特徵挖掘: 透過挖掘復發突變特徵 (RMS) 和染色質重塑突變特徵 (CRMS),有效預測
頭頸癌患者在接受治療後的復發風險。
3.多變數風險預測模型的建立: 基於淋巴結外侵犯、CCRT 反應、以及三種體細胞突變情形(TMB、RMS和
CRMS)所找出的預測性診斷模型,有效預測接受同步放射線及化學治療的頭頸癌病患的無復發生存期
【研究團隊】
團隊成員:王慧晶,魏芯樺,曾良鵬,吳俊杰,杜政勳,劉佩伶,周孟君,吳哲維,黃志仁,蕭惠樺,潘美仁,陳立宗
代表單位:醫學院、大數據研究中心
團隊簡介:本中心整合高醫大體系醫院與校院相關領域的菁英教授、專家,包括高雄醫學大學健康科學院(公衛系、醫管資系、醫放系等)、口腔醫學院(口衛系)、藥學院、醫學院等,並與高醫大醫療體系資訊室、醫學統計分析及生物資訊研究室、醫學資訊與統計中心等相關單位密切合作。
研究聯繫Email:joellewang66@gmail.com
【論文資訊】
論文出處:EXPERIMENTAL AND MOLECULAR MEDICINE v.55 n.5 p.926 -938
全文下載:https://www.nature.com/articles/s12276-023-00984-4
Miracle of Genetic Analysis in Head and Neck Cancer Treatment: A New Milestone in Personalized Medicine
Miracle of Genetic Analysis in Head and Neck Cancer Treatment: A New Milestone in Personalized Medicine
Personalized genetic profiling has emerged as a crucial approach in healthcare, aiming to enhance the effectiveness of treatments and predict the likelihood of developing specific health risks. The focus of this approach lies in identifying mutated genes through genetic testing and tailoring treatment plans accordingly. In the context of head and neck squamous cell carcinoma (HNSC), we sought to evaluate the significance of genetic profiling and tumor mutation burden (TMB) through advanced techniques like next-generation sequencing.
Our investigation honed in on the examination of two specific mutation signatures—relapse mutation signature (RMS) and chromatin remodeling mutation signature (CRMS). These signatures were explored to gain insights into predicting the risk of relapse in individuals undergoing treatment for HNSC, particularly those receiving concurrent chemoradiotherapy (CCRT) with platinum-based chemotherapy.
The study revealed noteworthy findings. Patients categorized into the high RMS and CRMS groups exhibited considerably shorter relapse-free survival compared to their counterparts in the low RMS and CRMS groups (p < 0.001 and p = 0.006, respectively). This underscores the potential significance of these mutation signatures in influencing treatment outcomes and relapse risks.
Further analysis using multivariate Cox regression delved into identifying independent risk predictors for HNSC relapse. Notably, factors such as extranodal extension, CCRT response, and three somatic mutation profiles—TMB, RMS, and CRMS—emerged as independent risk predictors. This comprehensive examination highlighted the intricate interplay of various factors influencing the relapse risk in individuals with HNSC.
To provide a practical application of our findings, we developed a predictive nomogram. This tool demonstrated commendable performance in forecasting relapse-free survival among patients with HNSC undergoing CCRT. By incorporating key variables such as extranodal extension, CCRT response, and the three somatic mutation profiles, the nomogram offers a valuable resource for clinicians in making informed decisions about treatment strategies and predicting the potential for relapse in individual cases.
In conclusion, our study underscores the pivotal role of personalized genetic profiling, specifically focusing on mutation signatures like RMS and CRMS, in predicting relapse risks for individuals with HNSC undergoing CCRT. The integration of these genetic insights alongside traditional clinical factors enhances our ability to identify patients at higher risk of relapse, facilitating more tailored and effective treatment strategies. The predictive nomogram further serves as a practical tool for clinicians, offering a comprehensive and personalized approach to managing HNSC and improving patient outcomes.
Graphical Abstract




Application and Highlights:
1.Next-Gen Sequencing for Comprehensive Genetic Analysis and TMB Assessment: Using advanced
sequencing, we gain a deeper understanding of \patients' genetic variations.
2.Predicting Relapse Risk through Mutational Signatures: Examining relapse mutation signatures
(RMS) and chromatin remodeling mutationsignatures (CRMS) effectively predicts relapse risk in head
and neck cancer patients post-treatment.
3.Multifactorial Risk Prediction Model: Considering extranodal extension, CCRT response, and three
somatic mutations (TMB, RMS, and CRMS), apredictive model forecasts relapse-free survival in head
and neck cancer patients receiving concurrent chemoradiotherapy.
Research Team Members:
Hui-Ching Wang, Sin-Hua Moi, Leong-Perng Chan, Chun-Chieh Wu, Jeng-Shiun Du, Pei-Lin Liu, Meng-Chun Chou, Che-Wei Wu, Chih-Jen Huang, Hui-Hua Hsiao, Mei-Ren Pan, Li-Tzong Chen
Representative Department: Medical School, Big Data Research Center
Introduction of Research Team:
The center integrates elite professors and experts from various disciplines within the Kaohsiung Medical University system, including the College of Health Sciences (Public Health, Health Care Management, Medical Informatics), College of Dental Medicine (Dental Hygiene), College of Pharmacy, and College of Medicine. Collaborative efforts extend to the Medical Information Department, Medical Statistics Analysis and Bioinformatics Laboratory, and the Medical Information and Statistics Center within the healthcare system of Kaohsiung Medical University.
Contact Email: joellewang66@gmail.com
Publication: EXPERIMENTAL AND MOLECULAR MEDICINE v.55 n.5 p.926 -938
Full-Text Article: https://www.nature.com/articles/s12276-023-00984-4






